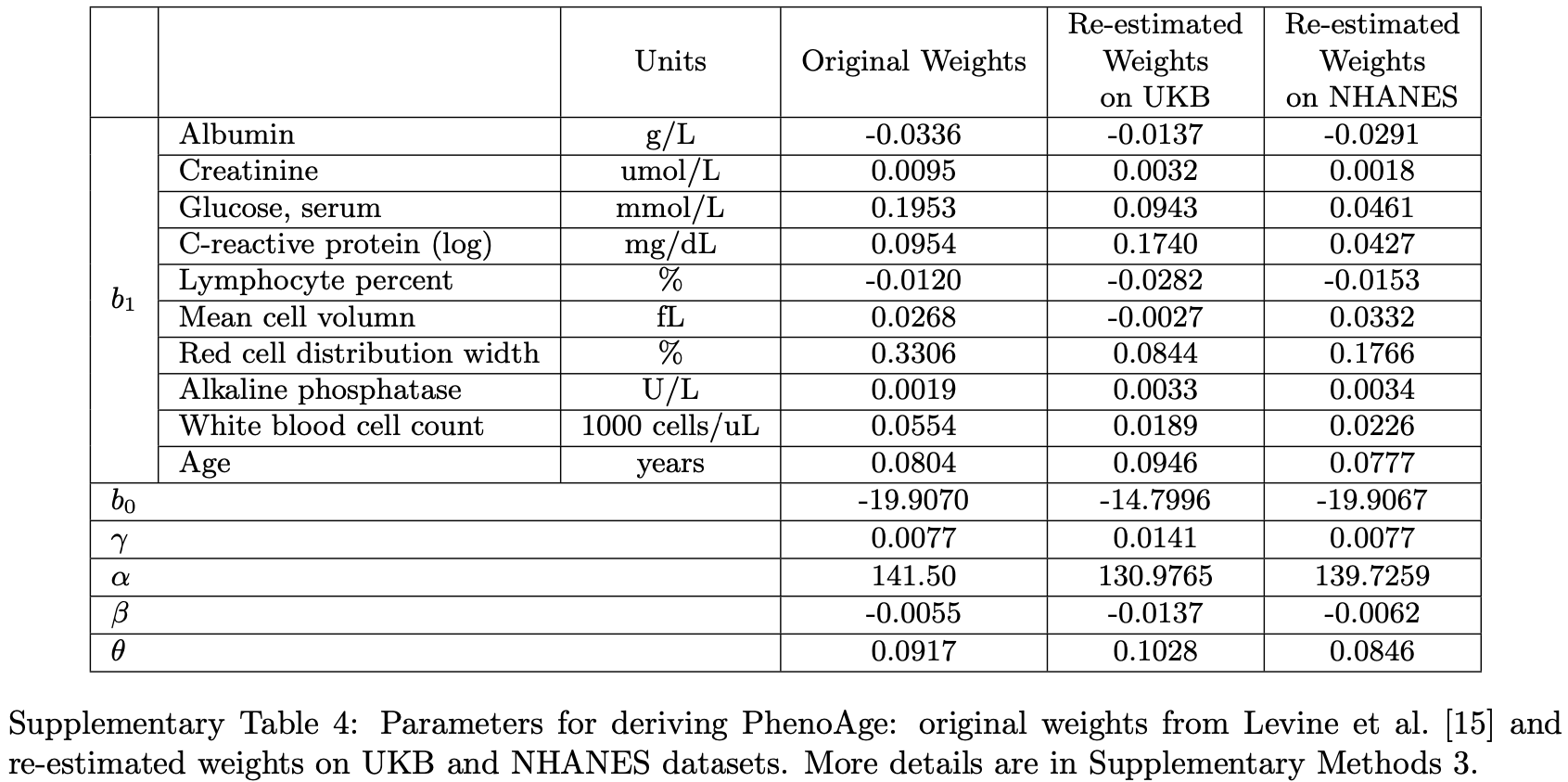
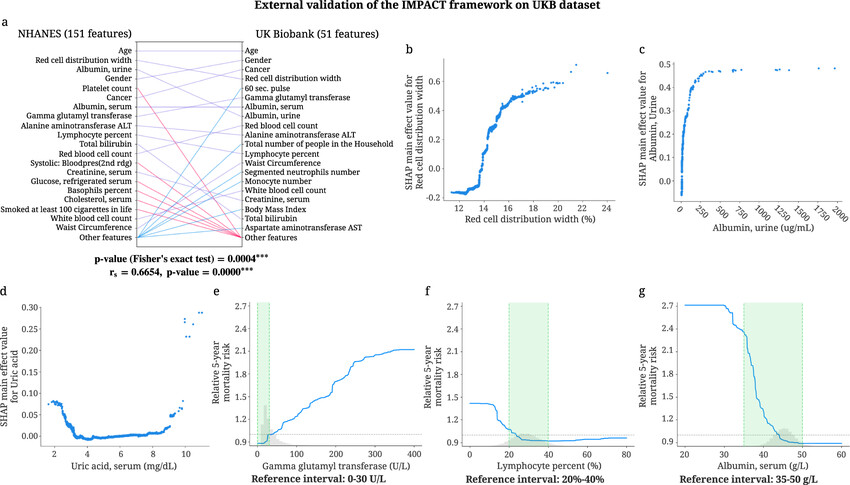
Here’s a study exploring the nationality/racial (Asian, Hispanic) variability in ranking mortality predictors:
" Here, we determine the strongest predictors of five-year mortality in four national, prospective studies of older adults. We analyze nationally representative surveys of older adults in four countries with similar levels of life expectancy: England (n = 6113, ages 52+), the US (n = 2023, ages 50+), Costa Rica (n = 2694, ages 60+), and Taiwan (n = 1032, ages 53+). Each survey includes a broad set of demographic, social, health, and biological variables that have been shown previously to predict mortality. We rank 57 predictors, 25 of which are available in all four countries, net of age and sex."
“C-reactive protein, additional inflammatory markers, homocysteine, serum albumin, three performance assessments (gait speed, grip strength, and chair stands), and exercise frequency also discriminate well between decedents and survivors when these measures are available. We identify several promising candidates that could improve mortality prediction for both population-based and clinical populations.”
“Laymen, social scientists, and health researchers have long shared a fascination with predicting a person’s longevity or future age at death. In a modern day version of fortune telling, the internet is replete with websites and “apps,” with such evocative names as “Death Clock,” “Death Timer,” and “Deadline,” designed to predict age at death or time remaining before death on the basis of a battery of simple questions.”
“In this paper, we use the extensive information collected in four recent biosocial surveys—in England, the US, Costa Rica and Taiwan—each of which is based on a large, national sample. We identify the best predictors of survival and assess robustness of these findings with multiple measures of discrimination. We address two specific questions. First, what factors matter most in determining whether an older adult will die in the next five years? Second, are the principal predictors the same across countries with similar levels of life expectancy but distinct social environments, levels of affluence, and access to health care?”
“One biomarker, C-reactive protein (CRP), is among the top 10 predictors in all four countries. Among the eight other biomarkers, only two appear among the top 10: glycosylated hemoglobin in Costa Rica and diastolic blood pressure in Taiwan. Self-reports of diagnosed heart disease, stroke, diabetes, and cancer appear on the list for some countries, but only in Taiwan does one of these (diabetes) appear among the five strongest predictors.”
“Peak expiratory flow (PEF), timed chair stands, and gait speed yield ΔAUC > = 0.01 among the countries in which they are available. Grip strength is a strong contributor in Costa Rica and Taiwan.”
“Several biomarkers perform very well among all countries for which they are available: serum albumin; homocysteine; and several inflammatory markers (fibrinogen, s-ICAM-1 and sE-selectin).”
“In England, the most notable differences are that the measures of cognitive function and grip strength are stronger predictors, while CRP is a weaker predictor in the full sample than in the restricted sample. Among the Taiwanese, two of the biggest changes involve education (which becomes a stronger predictor) and history of diabetes (which becomes a weaker predictor) when models are re-estimated for all interviewed respondents.”
“Nevertheless, exercise frequency appears on the top 10 lists for three countries, sometimes above measures of disease prevalence and biomarkers. This finding raises an important caveat, namely that our analysis cannot be used to infer causal linkages between the predictors and survival. For example, exercise may discriminate well partly because of reverse causality (poor health leads to reduced exercise).A major strength of this analysis is that we bridge the division between clinicians and researchers interested in mortality prediction and employ a more diverse set of predictors than previous studies.”
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4951106/