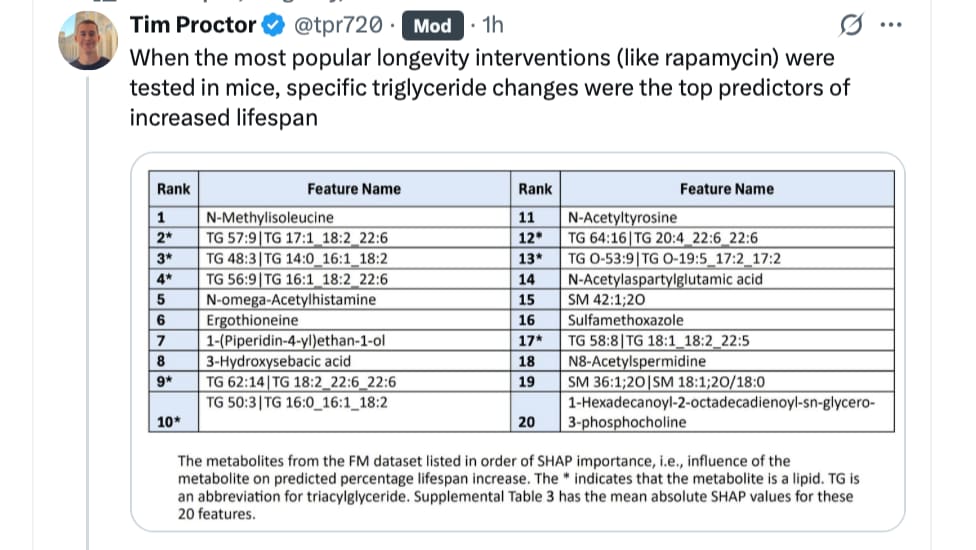
Researchers have developed a biological clock based on blood biomarkers that can accurately predict risk of mortality and frailty in mice. The findings, published in Nature Aging , lay the foundation for human biological clocks that health care providers can use to better understand how aging impacts patients differently.
This is interesting, and its out of Richard Miller’s lab (of the ITP fame).
Discrimination of Normal from Slow-Aging Mice by Plasma Metabolomic and Proteomic Features
Bretton Badenoch, Oliver Fiehn, Noa Rappaport, Pranjal Srivastava, Kengo Watanabe, Sriram Chandrasekaran and Richard A Miller
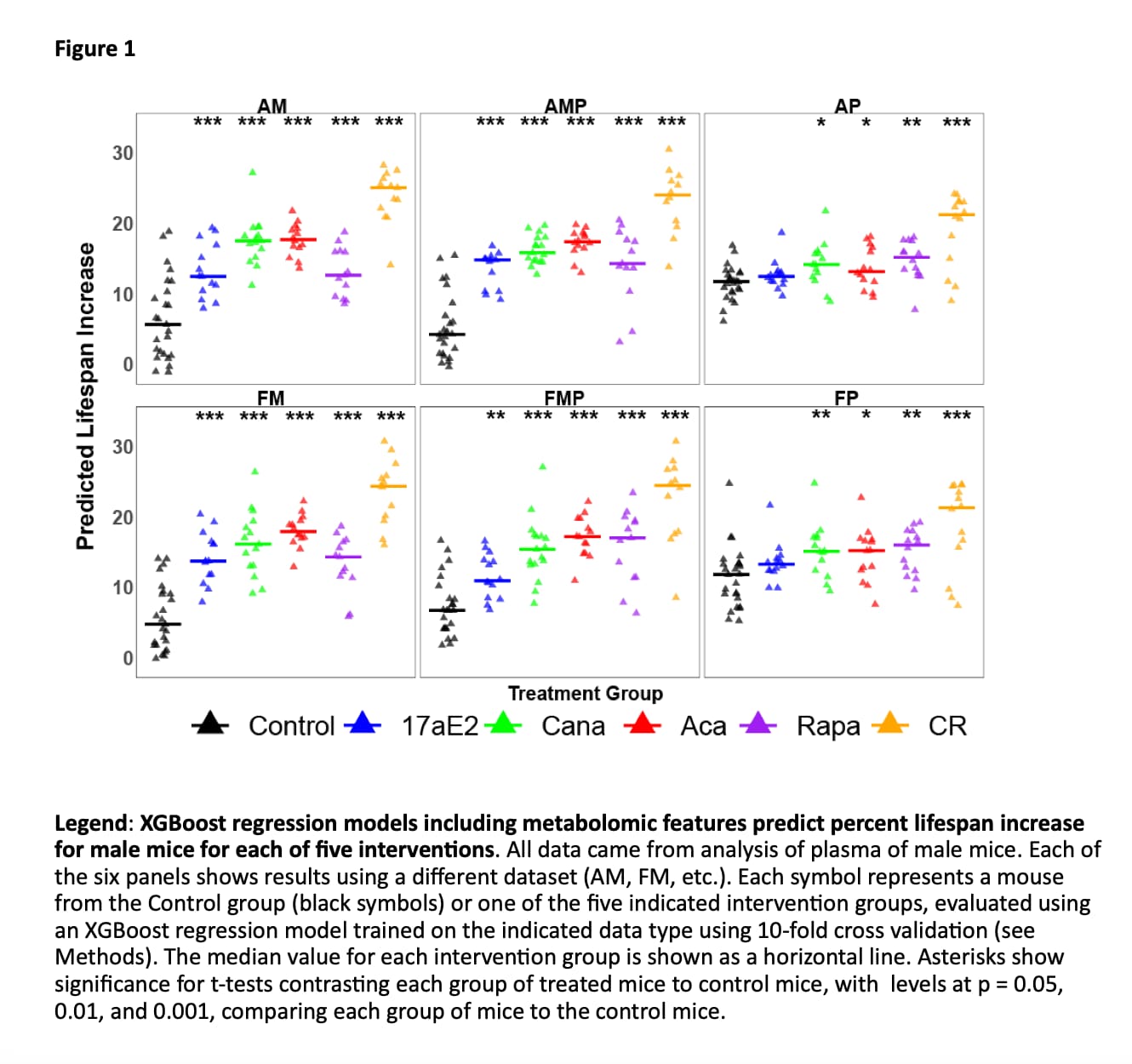
Tests that can predict whether a drug is likely to extend mouse lifespan could speed up the search for anti-aging drugs. We have applied a machine learning algorithm, XGBoost regression, to seek sets of plasma metabolites that can discriminate control mice from mice treated with an anti-aging diet (caloric restriction) or any of four anti-aging drugs. When the model is trained on any four of these five interventions, it predicts significantly higher lifespan extension in mice exposed to the intervention which was not included in the training set. Plasma peptide data sets also succeed at this task. Models trained on drug-treated normal mice also discriminate long-lived mutant mice from their respective controls, and models trained on males can discriminate drug-treated from control females. Triglycerides are over-represented among the most influential features in the regression models. Triglycerides with longer fatty acid chains tend to be higher in the slow-aging mice, while triglycerides with shorter fatty acid chains tend to decrease. Plasma metabolite patterns may help to select the most promising anti-aging drugs in mice or in humans, and may give new leads into physiological and enzymatic targets relevant to discovery of new anti-aging drugs.
@DrFraser perhaps some biomarkers we should be tracking while using rapamycin? Are we “hiding” these biomarker changes with lipid therapies like statins, bempadoic acid, Zetia, etc.?
Of the 20 metabolites that had the greatest influence on the estimation of lifespan predictions (FM
dataset), 8/20 (40%) were triacylglycerides (TG), although TG made up only 16% of the metabolites in this dataset (p = 0.003). Among the 20 highest-ranked TG (see Supplemental Table 2), FA with 20 – 22 carbon chains were most often at higher levels in the long-lived mice, while FA with 14 – 18 carbon chains were typically down-regulated. Interestingly, studies in model organisms have demonstrated that manipulation of lipid metabolism can influence lifespan. For example, overexpression of fatty acid desaturase genes, leading to increased production of unsaturated fatty acids like DHA, has been associated with extended lifespan in nematodes (30).
2025.05.11.651908v1.full.pdf (949.6 KB)
A few points - mouse mortality is greatly related to cancer, not vascular disease - the reverse is the case in humans. It’s complicated to move across this difference when looking at all cause mortality.
These markers are not commercially available at any of the labs I use (and I use over 40 labs).
I think I’d want some human data on this before trying to find a lab that would do these tests, and who knows what it would cost.
On a practical basis, I don’t see how I’d implement this in practice. I also have a long list of things that I am tracking, and another 20 unless there was some reasonable human evidence would have me reluctant to take time and my patient’s money.
It is an area to watch for sure, and it is fascinating research.
Can anybody access and share the full paper? The abstract says platelets were the best correlated.
Definitely interesting - but metabolomics is nowhere near as easy to implement as the usual biomarkers that we’re measuring. In my research, I’ve used metabolomics to profile cell culture medium and mouse plasma before, but for humans I’m not sure if we even have reference ranges. And I reckon the variability would be absolutely massive - all those different diets, microbiomes, medications etc. You’d need some absolutely massive “atlas” type study of the human metabolome to form a foundation. Maybe there’s already that sort of data in here: Molecular Transducers of Physical Activity Consortium (MoTrPAC) but I wouldn’t know how to use it.
Either way, definitely exciting times.