All the other epigenetic clocks have their issues (the 1st and 2nd generation clicks do not respond to calorie restriction, and even DunedinPACE increases after partial reprogramming). The meaningfulness of any of the CpG sites is unknown (and are not significant to the calculation of DamageAgea and AdaptAge). But the DamageAge and AdaptAge Ying CpG sites are different and the real ones
https://www.researchgate.net/publication/368982451_ClockBase_a_comprehensive_platform_for_biological_age_profiling_in_human_and_mouse
https://www.researchgate.net/publication/364266802_Causal_Epigenetic_Age_Uncouples_Damage_and_Adaptation
4 Likes
I got my data plugged in and there are VERY VERY sharp discrepancies that I don’t know what to make of yet…
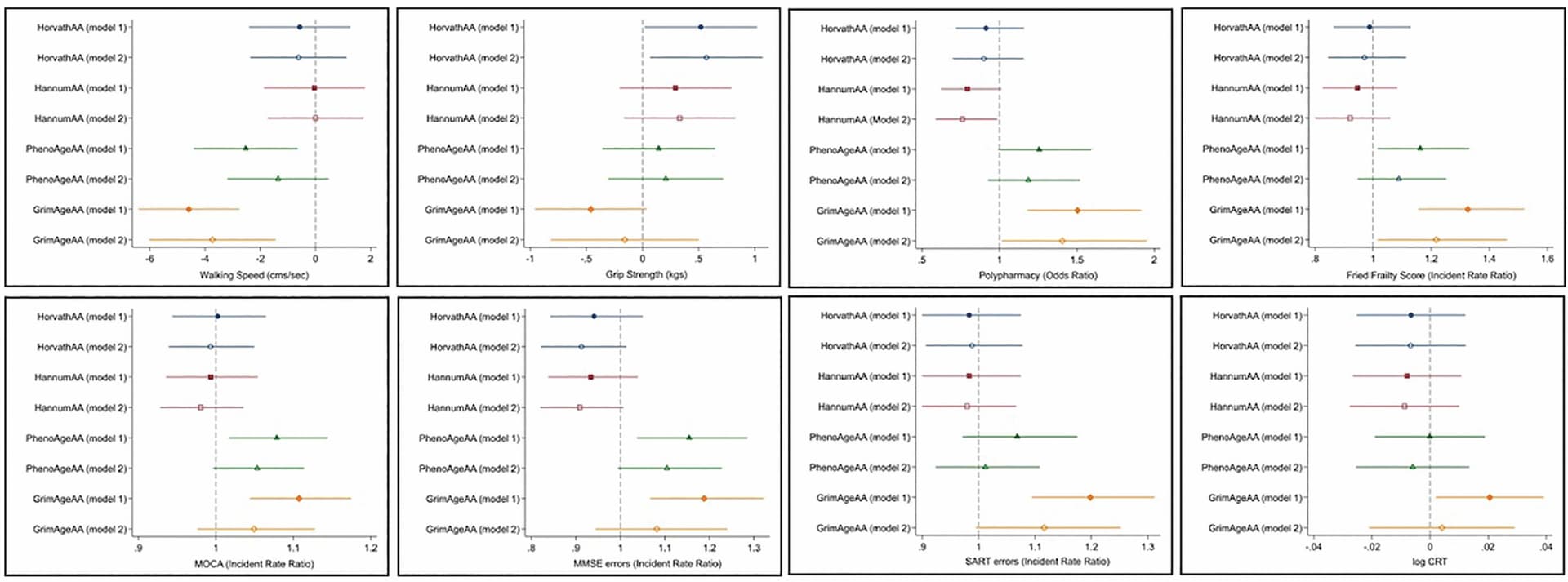
DunedinPACE is 0.59. Of all the epigenetic clocks, PhenoAge is by far the lowest (the first-generation epigenetic clocks have little functional significance and I can cry less over them b/c I do way better on 2nd/3rd generation). The first-generation epigenetic clocks are MORE correlated with AdaptAge than DamageAge (meaning HorvathAge and especially HannumAge are more associated with “adaptive/protective” changes w/age than damaging changes w/age).
But DunedinPoAM (precursor and less accurate than DunedinPACE) is barely over 1.
DamageAge and AdaptAge… I don’t know what to make of yet… There’s a huge discrepancy I need to figure out better, because there’s a chance this massive discrepancy could be rare enough to be socially important…
1 Like
HannumAge is MORE associated with adapt-age than damage-age, as is HorvathAge. DNAm that contribute to HorvathAge/HannumAge have little functional significance (in fact, slight acceleration of HannumAge may be protective - HannumAgeAA has the opposite effects of GrimAgeAA)
GrimAge outperforms phenoage…
Then there’s GrimAge2, which supposedly outperforms GrimAge (GrimAge2 is ONLY trained on people over 40 )
Talked to jesse pogalnik yesterday, who has some experience with it (and who recently published the DamageAge/AdaptAge papers). There’s thought that AdaptAge increases after recovery from stress [stress is when DamageAge may increase]. We still don’t know if DamageAge/AdaptAge are more dynamic/changeable/intervention-friendly than other clocks
1 Like
Kejun Ying has smg new
Upon adjusting for the number of genes in pathways, we found that the pathways
115 most predictive of aging in humans were primarily associated with lipid metabolism (Figure 2a).
116 The top five lipid metabolism pathways were: Linoleic acid metabolism (residual -1.51), Synthe-
117 sis of very long chain fatty acyl CoAs (residual -1.27), Alpha-linolenic (omega 3) and linoleic
118 (omega 6) acid metabolism (residual -1.16), Biosynthesis of unsaturated fatty acids (residual -
119 1.13), and Fatty acyl CoA biosynthesis (residual -1.04). Conversely, pathways that were least
120 predictive of aging (those with positive residuals) were mainly related to immune system regula-
121 tion and DNA replication. These include the Activation of C3 and C5 (residual 3.11), Assembly
122 of the ORC complex at the origin of replication (residual 2.64), and E2F-enabled inhibition of
123 pre-replication complex formation (residual 2.34)
ESR-mediated signaling and signal-
163 ing by nuclear receptors show a significantly slower or no decline in methylation rates with
in-
164 creasing lifespan, suggesting that these pathways may play conserved roles in aging processes
165 across mammals, regardless of species-specific longevity. In contrast, mRNA splicing showed
166 more rapid declines in methylation rates with increasing lifespan, hinting at potential
adaptive
167 changes in longer-lived species, possibly reflecting more efficient regulation of this
process in
168 animals with extended lifespans.
1 Like