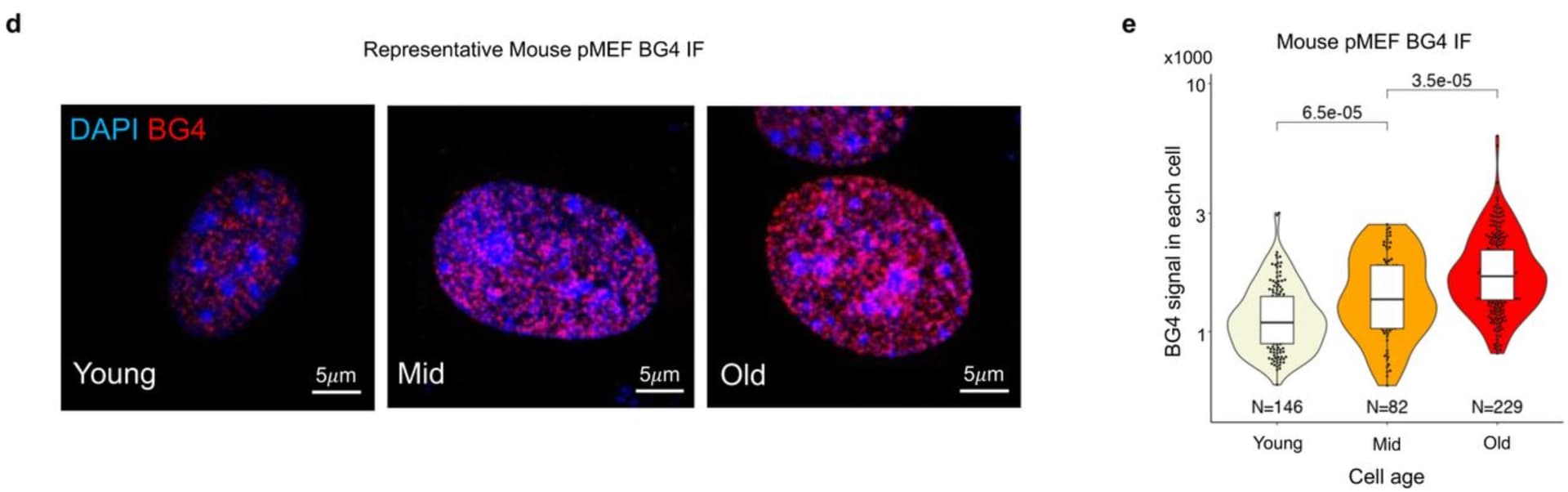
I don’t yet completely understand this paper, but one thing that stood out to me is that the BG4 immunofluorescence assay might be used to assess the efficacy of potential rejuvenating compounds. This assay allows you to fluorescently label DNA G-quadruplexes (G4s), the presence of which increases with cell passage, and which are linked to various aging markers (chromatin opening and impaired transmission of DNA methylation and histone modification) in this paper.
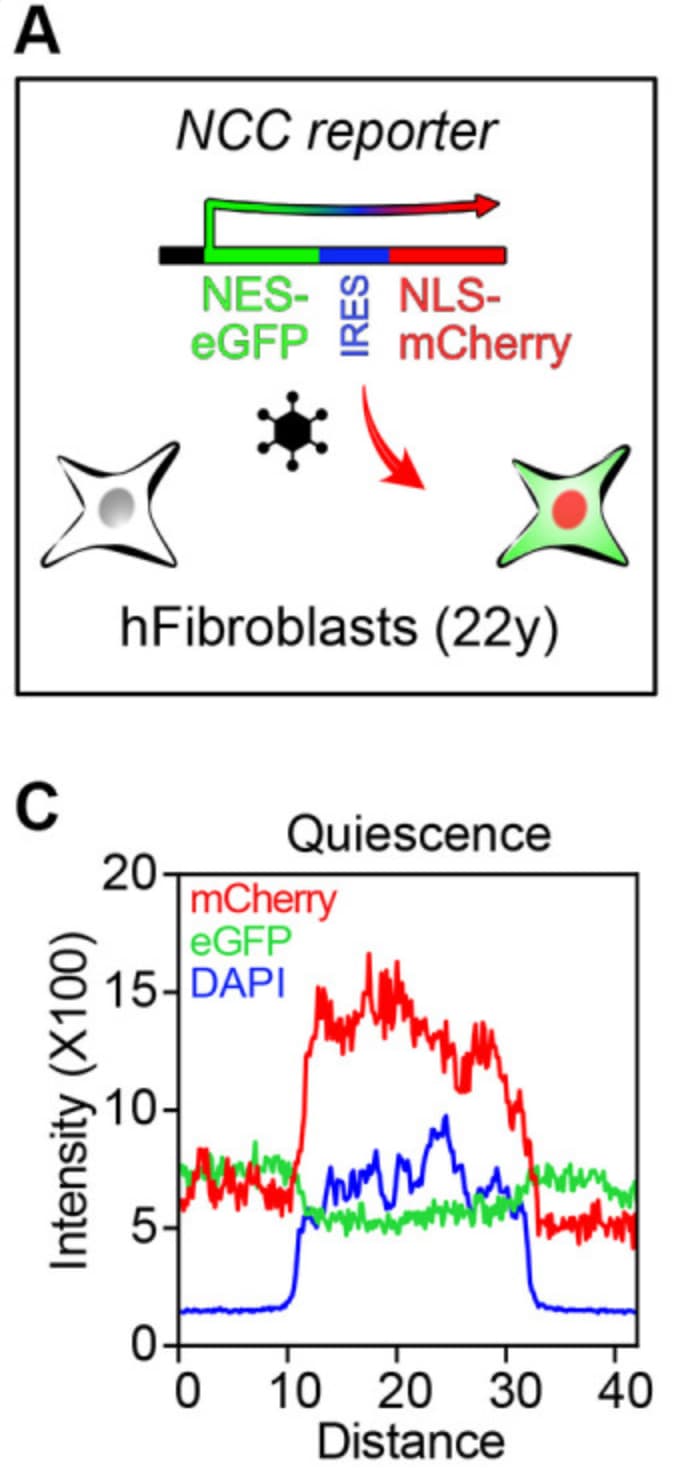
This could be used in tandem with the Sinclair lab’s nucleocytoplasmic compartmentalization assay for rejuvenating compounds. In aging, nuclear proteins are improperly localized to the cytosol, and rejuvenating compounds may reverse these changes.
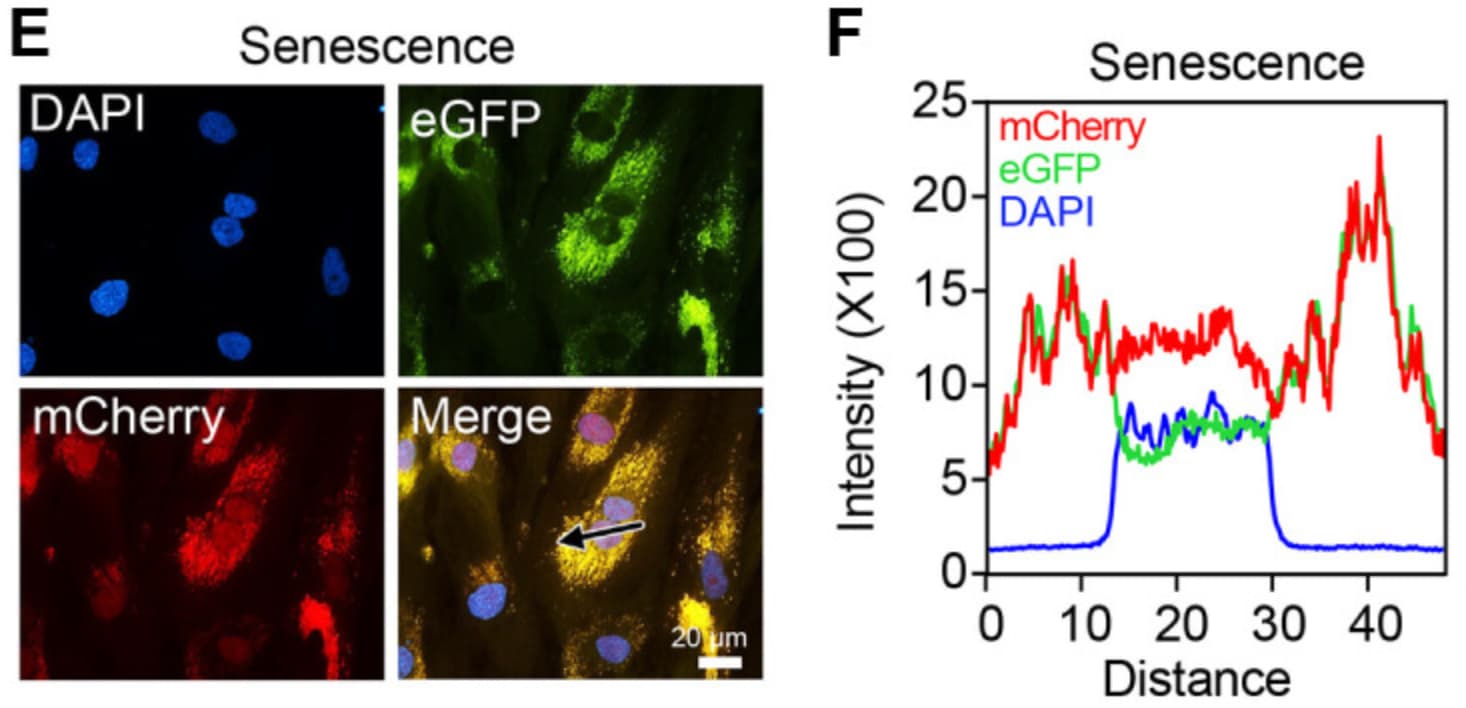
You can see below that in the quiescent fibroblasts, mCherry is targeted to the nucleus (note the overlap with the nuclear marker DAPI) due to its nuclear localization signal.
Whereas below there is significant loss of nuclear mCherry, due to replicative senescence being induced in the fibroblasts.
Also, this paper was focused on aging mechanisms related to cell division, so it especially relates to aging of tissues or structures (gut, skin, hippocampus, etc) that are replenished throughout life.
The authors mention that whether G4s contribute to aging of non-mitotic cells is an open question.
Finally, our work focused on the G4 formation cycle throughout mitosis. However, aging also occurs in cells that do not actively divide, such as neurons. The chromatin accessibility of neurons shifts during aging114, suggesting that erosion of the epigenetic landscape might also underlie aging in these cells. However, whether G4s might actively regulate aging in these non-dividing cells, and if so, what the underlying molecular mechanism is, is an open question awaiting future investigation.
One emerging mechanism of aging is increased stalling of RNA polymerases which disproportionally affects the transcription of longer genes. It’s tempting to speculate whether intragenic G4 formation might impede RNA polymerase elongation and partially contribute to the above finding.
We found that in 2-year-old liver, 40% of elongating RNA polymerases are stalled, lowering productive transcription and skewing transcriptional output in a gene-length-dependent fashion. We demonstrate that this transcriptional stress is caused by endogenous DNA damage and explains the majority of gene expression changes in aging in most mainly postmitotic organs, specifically affecting aging hallmark pathways such as nutrient sensing, autophagy, proteostasis, energy metabolism, immune function and cellular stress resilience. Age-related transcriptional stress is evolutionary conserved from nematodes to humans. [ref]
Just reading the abstract it seems to imply hypomethylation (of DNA) when AIUI aging is associated wth more methylation not less.
Global hypomethylation but CpG islands often are hypermethylated in aging.
I thought it was the CpG islands that are normally measured by methylation clocks.
See 2013 paper DNA methylation age of human tissues and cell types by Horvath
The 353 clock CpGs can be divided into two sets according to their correlation with age. The 193 positively and 160 negatively correlated CpGs get hypermethylated and hypomethylated with age, respectively
…
While positively related markers do not show a significant relationship with CpG island status (Additional file 9F), negatively related markers tend to be over-represented in CpG shores (P = 9.3E-6; Additional file 9K).
Perhaps in more recent clocks that have been developed, the hypermethylated clock CpGs are over-represented in CpG islands, but I’m not familiar with the literature on that.