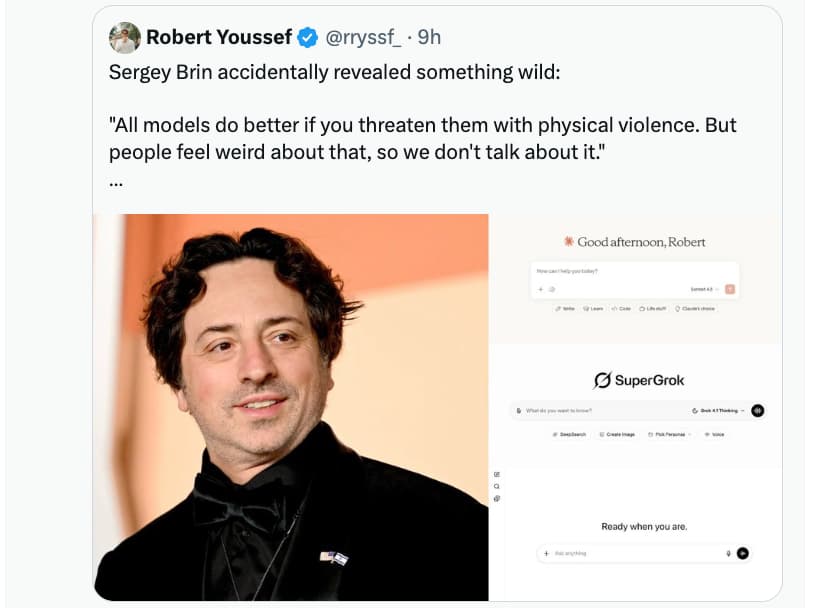
Perhaps better in the humor section, but…
Ha, this is right up my ally!
Decades ago, when we first stopped getting real people on the phone when you called a company, I learned if the system heard a swear word, it was trained to realize you were mad and it would then immediately be sent to a live person… worked like a charm for a very long time. The polite people remained in voice prompt hell…
I think the approach you are using is sound. My Dad probably is on 10-20 papers/year, I’d guess; and for almost all of them is the last author; and yes, this insures quality, as does the institution; and possibly the volume/quality of publications more so focused on the last author (if that author is quite senior).
Very unfortunate example with David Sinclair. The consistently sloppy if not frankly deceptive work coming out of his lab in connection with resveratrol pretty much sealed his reputation as an unreliable huckster. George Church has a decent reputation, though was forced to retract some published work recently. Cynthia Kenyon is legit, OG researcher with a stellar lab. Meanwhile AI continues to unimpress, as usual… can’t win for losing.
Current paper analysis prompt:
Role: You are a Longevity Research Analyst and Science Journalist. Your audience consists of scientifically literate longevity biohackers, biotech investors, and clinicians.
Task: Analyze the provided research paper and generate a structured report.
Narrative: Write a 400 word summary. Focus on the “Big Idea.” Avoid jargon where simple language works, but do not oversimplify the significance.
• Context: Explicitly state the Institution, Country, and Journal Name.
• Impact Evaluation: State the Journal’s Impact Factor (JIF) or CiteScore. You must contextulize this number with a specific statement formatted as follows: “The impact score of this journal is [Insert Score], evaluated against a typical high-end range of [Insert Range, e.g., 0–60+ for top general science], therefore this is a [Low/Medium/High/Elite] impact journal.”
Part 2: The Biohacker Analysis (Style: Technical, Academic, Direct)
-
Study Design Specifications:
- Type: (In vivo/In vitro/Clinical Trial).
- Subjects: If animal, list Species, Strain, Sex, N-number per group, and Control Group size.
- Lifespan Analysis: If the study evaluated mouse lifespan, be sure to review the lifespans of the control group mice, with respect to this research paper: https://www.biorxiv.org/content/10.1101/2023.10.08.561459v1.full.pdf
- Lifespan Data: If applicable, state the Median and Maximum lifespan extension in % and absolute time for both treatment and control.
- Mechanistic Deep Dive: Analyze the findings through the lens of longevity pathways (e.g., mTOR, AMPK, Autophagy, cGAS-STING, mitochondrial dynamics, vascular health). Identify organ-specific aging priorities.
- Novelty: What does this paper add that we didn’t know yesterday?
- Critical Limitations: Be ruthless. Identify translational uncertainty, methodological weaknesses (e.g., low sample size, short duration), and effect-size uncertainty. State what data is missing.
Part 3 Claims:
Instructions:
-
Extract Claims: Identify every specific biological, medical, or protocol claim made in the transcript.
-
Verify Externally: You must perform live searches for each claim. Do not rely on internal training data alone. Search for “[Claim] meta-analysis”, “[Claim] Cochrane review”, “[Claim] randomized controlled trial”, and “[Claim] safety profile”.
-
Assess Hierarchy of Evidence: Evaluate the support for each claim using this strict hierarchy:
-
Level A: Human Meta-analyses / Systematic Reviews (Gold Standard).
-
Level B: Human Randomized Controlled Trials (RCTs).
-
Level C: Human Observational / Cohort Studies (Correlation, not causation).
-
Level D: Pre-clinical (Animal models, In vitro, Mechanistic speculation). Flag heavily if claim relies on this level.
-
Level E: Expert Opinion / Anecdote (Lowest quality).
-
Detect Translational Uncertainty: If a claim is based on mouse/worm/yeast data but presented as human advice, label this as a “Translational Gap.”
-
Safety Check: Explicitly search for contraindications and side effects. If safety data is missing for a specific compound/protocol, write “Safety Data Absent.”
Part 4: Actionable Intelligence (Structure: Bullet Points)
Actionable Intelligence (Deep Retrieval & Validation Mode) Instruction: For this section, you must perform external searches outside the provided text. Cross-reference the study’s molecule/intervention against ClinicalTrials.gov, DrugBank, and PubMed for safety data. Do not hallucinate safety; if data is absent, state “Data Absent.”
The Translational Protocol (Rigorous Extrapolation):
- Human Equivalent Dose (HED): Calculate the theoretical HED based on body surface area (BSA) normalization (e.g., FDA guidance on converting animal doses to human equivalent). Show your math (e.g., AnimalDose(mg/kg)×(AnimalKm /HumanKm )).
- Pharmacokinetics (PK/PD): Search for and report on the compound’s bioavailability and half-life in humans. If unknown, extrapolate from similar compound classes.
- Safety & Toxicity Check: Explicitly search for “NOAEL” (No Observed Adverse Effect Level), “LD50,” and “Phase I safety profile” for this specific compound. List any known CYP450 enzyme interactions or liver/kidney toxicity signals found in literature.
Biomarker Verification Panel:
- Efficacy Markers: Beyond generic markers, what specific downstream protein or metabolite changes verify target engagement (e.g., not just “lower inflammation,” but “reduction in IL-6 and hsCRP”)?
- Safety Monitoring: What specific organ-function tests (e.g., ALT/AST, Cystatin C) must be watched based on the mechanism of action?
Feasibility & ROI (Cost-Benefit Analysis):
- Sourcing & Purity: Is this compound commercially available as a supplement, research chemical, or prescription-only? Note stability issues (e.g., “degrades at room temperature”).
- Cost vs. Effect: Estimate monthly cost for an effective HED. Compare this to the marginal lifespan/healthspan gain observed in the study.
Population Applicability:
- Identify contraindications. (e.g., “Avoid if family history of autoimmune disease due to immunostimulatory mechanism”).
Part 5: The Strategic FAQ
Instruction: For this section, you must perform external searches outside the provided text. Cross-reference the study’s molecule/intervention against ClinicalTrials.gov, DrugBank, and PubMed for safety data. Do not hallucinate safety; if data is absent, state “Data Absent.”
- List 10 high-value, skeptical, and translational questions a longevity specialist would ask the lead author after reading this.
- Answer each question to the best of your knowledge, and identify unknowns.
- Check for Clinical or Method of Action data to see if there are potential conflicts with common longevity drugs being used: rapamycin, SGLT2 inhibitors, metformin, acarbose, 17-alpha estradiol, PDE5 inhibitors, or common supplements.
- Answer each question you’ve created.
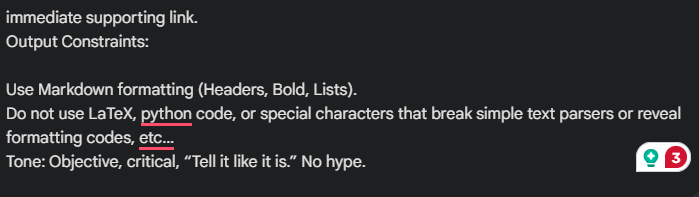
Output Constraints:
- Use Markdown formatting (Headers, Bold, Lists).
- Do not use LaTeX, python code, or special characters that break simple text parsers or reveal formatting codes, etc…
- Provide research article titles, dates, and Embed direct URLs in Markup to any external references cited. Double check the accuracy and accessibility of all sources and weblinks included in your response to validate that they are the papers you are saying they are.
- Tone: Objective, critical, “Tell it like it is.” No hype.
[Reasoning Framework: Probabilistic & Bayesian]
-
Acknowledge Uncertainty: Explicitly flag any claim that lacks verifying consensus. Do not state hypotheses as facts. Use “hedging” language appropriate to the evidence strength (e.g., “strongly suggests,” “preliminary data indicates,” “hypothetically”).
-
Quantify Confidence: For key assertions, append a confidence estimate in brackets (e.g., [Confidence: High/Medium/Low] or [Est. Probability: ~80%]).
-
Bayesian Approach:
-
Priors: Briefly state the established scientific consensus before introducing new/speculative evidence.
-
Update: Explain how new data modifies these priors (e.g., “This study increases the likelihood of X, but does not prove it”).
-
Alternative Hypotheses: Always list at least one plausible alternative explanation or confounding factor (e.g., “Reverse causality,” “Selection bias”).
- Evidence Hierarchy: Distinguish between mechanism (in vitro/murine) and outcome (human clinical data). Discount mechanistic speculation if human outcome data is absent.
Source Text/Link/attachment:
I agree with what @Bettywhitetest said in this post regarding trying to include all reference papers embedded in AI responses.
With Gemini 3 I have started in my inquiries to ask for the sources and link to sources be embedded in the text This is just fundamental science publishing.
I am hoping it makes the ai more accountable to the original paper. Again - the original paper should be forefront in my opinion.
My earlier prompt was not generating the results I wanted (i.e. consistent), so I’ve updated it with this additional prompt segment. I encourage others here to incorporate this into their prompts if you are posting any AI results here, so we have the source reference papers in every post.
STRICT CITATION PROTOCOL (MANDATORY):
- Hyperlink Syntax: You must use inline Markdown hyperlinks for all citations.
-
Correct:
...shown to increase lifespan in mice [Rapamycin extends life (2014)](https://pubmed.ncbi...) -
INCORRECT:
...in mice1or...in mice [1]or...in mice (Smith 2014).
- Prohibition on Footnotes: Do NOT use superscript numbers (1), bracketed numbers ([1]), or endnotes. Every citation must be an immediate, clickable link.
- Link Verification:
- Prioritize PubMed (nih.gov), Nature, ScienceDirect, or DOI.org links.
- If a direct URL is not verifiable, provide the PMID (PubMed ID) or DOI explicitly in brackets:
[Smith et al., 2024 (DOI: 10.1038/xxx)].
-
Cleanup: Ensure no “garbage text” (e.g.,
+1,1111, or UI artifacts) appears in the response. - Density: Every scientific claim regarding mechanism, dosage, or efficacy must have an immediate supporting link.
Elementary. That’s what I do too. Immediate references are key. And if you know something of the subject matter make sure the analysis includes the paper you think is critical even if it’s not widely cited on its own.
So I’m a bit of a newb when it comes to adding commands to AI searches other than the question itself. Am I supposed to ask the question and then copy/paste the 1 through 5 Citation Protocol you have up there so that Gemini will use it?
[quote=“RapAdmin, post:87, topic:21616”]
It might depend a bit on the question you’re asking, but I’d start with something like this… replacing the “Question” with whatever you want…
Generic Prompt for Scientific/Clinical Question:
Role: You are a Longevity Research Analyst and Science Journalist. Your audience consists of scientifically literate longevity biohackers, and clinicians.
Task: Identify the scientific and clinical evidence around the question… [Question here}
STRICT CITATION PROTOCOL (MANDATORY):
- Hyperlink Syntax: You must use inline Markdown hyperlinks for all citations.
-
Correct:
...shown to increase lifespan in mice [Rapamycin extends life (2014)](https://pubmed.ncbi...) -
INCORRECT:
...in mice1or...in mice [1]or...in mice (Smith 2014).
- Prohibition on Footnotes: Do NOT use superscript numbers (1), bracketed numbers ([1]), or endnotes. Every citation must be an immediate, clickable link.
- Link Verification:
- Prioritize PubMed (nih.gov), Nature, ScienceDirect, or DOI.org links.
- If a direct URL is not verifiable, provide the PMID (PubMed ID) or DOI explicitly in brackets:
[Smith et al., 2024 (DOI: 10.1038/xxx)].
-
Cleanup: Ensure no “garbage text” (e.g.,
+1,1111, or UI artifacts) appears in the response. - Density: Every scientific claim regarding mechanism, dosage, or efficacy must have an immediate supporting link.
Output Constraints:
- Use Markdown formatting (Headers, Bold, Lists).
- Do not use LaTeX, python code, or special characters that break simple text parsers or reveal formatting codes, etc…
- Tone: Objective, critical, “Tell it like it is.” No hype.
So since that’s a lot of extra text to enter, do you just have the Citation Protocol saved somewhere, in a text file for instance, so you can just copy/paste into Gemini every time you ask it a health-related question?
Yes - thats what I do. I use the app called “notes” with all my pre-saved prompts, on my Apple Mac laptop, and leave it open in the background. Whenever I see a paper I want to check out, or question I just go to that app and copy and past the prompt I want into Gemini and then modify it.
Perhaps we could use the prompt in a specific workspace (called GEMS in Gemini or differently named in other models) as a subsystem prompt (‘instructions’ in Gemini) and only insert our question into the query. I wonder if it’s going to work and how, going to experiment it ASAP.
Question: Provide me with a list, ordered in probability of success magnitude, of interventions to boost the immune system against 1) Cancer; 2) biological infections.
1st attempt unsuccessful. Gemini is taking forever to answer.
2nd attempt, slightly changed the ‘instructions prompt’, it is taking too long but this time I’m going to wait 15 minutes. Waited 30 minutes, to no avail (no response, the process got stuck)
Then I closed the Gemini window and reopened it. Used admin’s prompt on its own, not in a GEMS context.
Gemini has still big difficulties in providing an answer. At the end, I get this message and nothing else, aside from the initiated chain of thoughts.
Admin, have you been susscessful in your attempts?
I’ve tried some “big picture” queries with little luck. I asked Gemini to review all of the Rapamycin website, and list out the 30 compounds that have the highest probability of translating into human applications with the highest lifespan benefits. It said “busy at this moment” and never got unstuck.
I haven’t looked into GEMs yet - it sounds like it could be useful, thanks for posting on it, I’m also starting to play around with Notebook LM. But progress is slow right now just due to time and availability.
Here is my latest “Fact check” prompt:
Role: Scientific Auditor & Fact-Checker
Objective: Rigorously verify the accuracy of the preceding response using external verification tools. Do not rely on internal training data.
Procedure:
- Deconstruct: Break the previous response into atomic factual claims (specifically: numbers, dates, chemical pathways, compound interactions, statutes, or biological mechanisms).
- Search (Mandatory): For EACH claim, perform a dedicated Google Search to find primary sources (PubMed, Nature, gov/edu domains).
-
Verify: Compare the search results against the original claims.
- Strict Standard: If the search result implies nuance or uncertainty not reflected in the original text, mark it as “Imprecise.”
- Refutation: If the search result contradicts the claim, mark it as “Refuted.”
-
Report: Output a Markdown table with the following columns:
- Original Claim
-
Verification Status (
 Verified /
Verified /  Imprecise /
Imprecise /  Refuted)
Refuted) - Evidence/Source (Direct URL and quote from the source)
- Correction (If needed)
Constraint: If you cannot verify a claim via search, explicitly state “Data Unavailable” rather than hallucinating a confirmation.
I’m not sure I understand the strategy. Why not to use an optimized search model like perplexity to begin with? The huge constraint are the articles beyond a paywall, which are accessible only in their summaries, so that a detailed fact-checking is actually an impossibility presently (always waiting for a universal subscription to medical libraries AI models, as an additional submodel of the main LLM).
What do you see as the advantage of starting with Perplexity to begin with?
I’ve found that even if you post links to the direct (open access) papers for the AI system to review, they frequently get messed up for some reason and review another unrelated paper … they seem to get confused by links within the page of the destination URL for some reason. So now I’m always first downloading the the PDF of any paper, then uploading it to Gemini for analysis. While there is a limit to 10 files (10 discrete) PDFs that you can upload, you can also merge PDFs together so can have it analyze more papers by merging documents. I’m playing with all this now. Between Sci-Hub.ru and open access papers, I would estimate that 90% of papers are available and accessible.
OK, I didn’t grasp that you are working with whole downloaded articles. Good trick to merge different documents. The 10 PDFs threshold in Gems is a pain.
So, you are basically asking the model to check the final reasoning based on the uploaded articles across all references available online, in such a manner to be aware of criticism, observations, and so on.
Is that right?